This is another part of my blog series on molecular modelling. I use GAMESS for the QM calculations, and Avogadro and wxMacMolPlt as the GUI.
This post is going to be about basis sets, mainly about their usage in GAMESS, with a little bit of theory at the beginning. Skip the first three points if you don’t want that.
Orbital Approximation
In QM calculations, we attempt to solve the Schrödinger Equation for chemical systems (molecules, ions etc). If we could solve it exactly, then we would have the wavefunction for the system, which would give use all the properties (including energy) of the system. Unfortunately, it is not possible to have analytic solutions for systems larger than a hydrogen atom, so a lot of approximations are used to get something that can be solved. And guess what! even after all of those approximations, we can still get very very good match between calculated and experimental results.
Anyway, in case of chemical species, one approximation used is the orbital approximation, which assumes that we can separate the wavefunction of all electrons into components called orbitals (each of which can hold a maximum of two electrons). In atoms these are called atomic orbitals(AOs), in molecules (or any multi-atom system), molecular orbitals (MOs).
Basis sets
As chemists, we mostly deal with molecular orbitals. In QM softwares, MOs are usually expressed as linear combinations of the AOs of the atoms that make up the molecule. This is the Linear Combination of Atomic Orbital approach (LCAO).
That means we need the functions of the atomic orbitals first. We can use the orbitals of the hydrogen atoms (which can be found analytically) as guess. But these orbitals contain an e^(-kr) term (r is the distance from the nucleus, k is a constant), and calculating integrals (which is a step in the calculation) with these orbitals is computationally expensive. An easy solution is to use Gaussian type functions instead, which have the term e^(-kr²). Integrals are much faster with these. But they are not atomic orbitals! So what we do is that we express one atomic orbital as a linear combination of multiple Gaussian type functions.
So, we express the all atomic orbitals of any atom with a set of Gaussian type functions. This is called the basis set. (Physicists may sometimes use plane wave basis sets for calculations on crystals or periodic systems, but chemists rarely use them)
Polarisation and diffuse functions
One problem is that the hydrogenic atomic orbitals can only describe a free non-bonded atom (e.g. s-orbitals are perfectly spherical). But in a molecule, the orbitals of an atom may get polarized (e.g. an s-orbital might get elongated) by the presence of surrounding atoms. To correct for this error additional functions are added (e.g. a p-type function to polarize an s-type function), these are called polarisation functions and are often necessary to describe bonding.
Another problem is that for anions the orbitals can extend very far away from the atom. The only way to do this is to change the constant in the exponent, k, but this is usually fixed from the start in a software. So, some extra functions with very small exponents are added, so that accurate representation of the behaviour of electrons can be modelled.
Common basis set types
The Gaussian functions that I mentioned earlier have a constant at the front and an exponent constant k. Various researchers have found various combinations of these two constants that are suited for different types of calculations. This is why there is a wide range of basis set types of to choose from. Some common basis set types (that are all available in GAMESS) are:
1.Pople type basis sets:
These are quite old, they were made mainly for Hartree-Fock type calculations, but they are still popular for DFT calculations as well. They were proposed by the famous theoretical chemist John Pople (he won a Nobel Prize!). An example is the 6–311++G** basis set. The first number (6) means that each inner orbital is expressed with 6 Gaussian functions. The three numbers after the hyphen (3,1,1) means that the valence orbitals are split into 3 orbital functions each with 3,1 and 1 Gaussian functions respectively. The + symbols mean diffuse functions added (two + means diffuse functions added to both heavy and hydrogen atoms). Finally the * represents polarisation functions (so polarisation functions added to both heavy and hydrogen atoms). It is also sometimes written as 6–311++G(d,p). There are also double split valence basis sets like 6–31+G*.
Choosing them in GAMESS is a bit difficult. If the basis set is x-yz type then use GBASIS=Nyz and NGAUSS=x. So for 6–31G it would be GBASIS=N31 and NGAUSS=6. For 6–311G it would be GBASIS=N311 and NGAUSS=6. For adding polarisation functions use NDFUNC (for heavy atoms), NPFUNC (for hydrogens) and NFFUNC. For diffuse functions use DIFFSP=.t. (for heavy atoms) and DIFFS=.t. (for hydrogens). (Better option is to use the Avogadro input builder)
2. Jensen type basis sets:
These are also called polarisation-consistent basis sets (proposed by Frank Jensen). They are represented as pc-0, pc-1, pc-2 ,… the number representing the level of polarisation function added. If you want diffuse functions, you have to use the augmented version (aug-pc-0, aug-pc-1,…).
These basis sets are relatively recent, and are optimized for both DFT and Hartree-Fock calculations.
To use them in GAMESS, use GBASIS=PCseg-0 (or PCseg-1 etc) and add ISPHER=1 in $CONTRL. For augmented versions, use GBASIS=APCseg-1 etc.
3. Dunning type basis sets:
These are also called correlation-consistent basis sets (invented by Thom Dunning). These are represented as cc-PVDZ, cc-PVTZ, cc-PVQZ, cc-PV5Z… like Jensen basis sets. Augmented versions are also available (aug-cc-PVDZ, aug-cc-PVTZ,…).
These basis sets are also recent and are optimized for post-Hartree Fock methods (such as MP2, CCSD(T) etc.).
To use them in GAMESS use GBASIS=CCD (or CCT, CCQ, CC5 etc.) and ISPHER=1 in $CONTRL. Augmented versions can be requested with GBASIS=ACCD etc.
4. Ahrichs type basis sets:
Also known as Karlsruhe basis sets, they were developed by Reinhart Ahlrichs. There are two versions of these, the older ones were named def-SVP, def-TZVP, whereas the newer versions are named def2-SVP, def2-TZVP, def2-SVPD, def2-TZVPP etc. (D means diffuse functions added) The newer versions have been found to be much more accurate, but they are not included in GAMESS right now. However, you can load them externally (I will come onto that later).
The basis sets with one ‘P’ at the end are made for DFT calculations, whereas those with ‘PP’ at the end are for post-HF. The advantage over Jensen type is that Ahlrichs basis sets are available for all elements from H to Rn (almost all of periodic table). However, some basis sets of this family can sometimes give wrong relative energies (see this paper).
Type of contraction?
Some basis sets have the same Gaussian function appear in multiple orbital functions. This is called general contraction. In other basis sets one Gaussian function appears only once. This is called segmented contraction.
You would think that the same Gaussian function appearing multiple times would simplify the calculation. Unfortunately, most QM programs (including GAMESS) cannot deal with general contraction, what they do instead, is redo the integrals for every orbital function (meaning the same integrals are computed multiple times, which is very inefficient). Segmented contractions are far easier to deal with, and therefore faster.
Pople, Jensen, and Karlsruhe type basis sets all have segmented contractions. (The original version of Jensen type basis set was generally contracted, but it was replaced by the segmented version in most programs, including GAMESS). On the other hand, Dunning type basis sets are generally contracted.
Getting external basis sets
One of the best places to get basis sets is the Basis Set Exchange library (https://www.basissetexchange.org/). Choose the basis set that you need on the left, then select the atoms that you need on the periodic table on the right (the elements available in that basis set will have a small yellow sticker at the bottom left, if you need other elements, then choose another basis set). Then select ‘GAMESS US’ in Format, and click Get Basis Set.
A new window will open, and you can then copy the text from it.
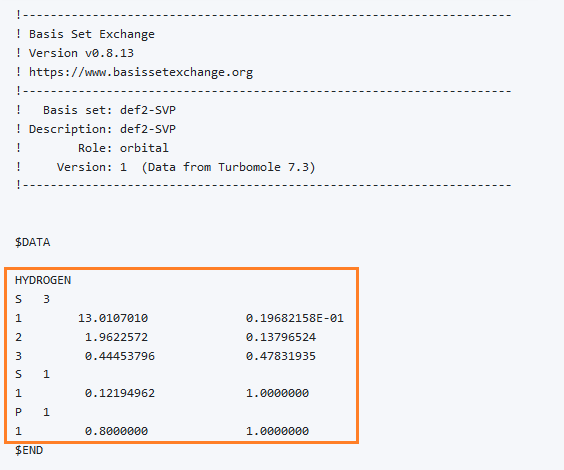
def2-SVP basis set for hydrogen
The exact meaning of those numbers does not matter right now.
Using external basis sets in GAMESS
Any basis set that is defined with Gaussian functions can be dealt with by GAMESS. There are multiple ways to load a basis set that is not available by default. I will go through them one by one.
- Inputting in DATA
If you are inputting the $DATA section in cartesian coordinates then it is possible to add the basis sets for each atom after the line defining their cartesian coordinates. For example, this the $DATA section for a hydrogen molecule, with def2-SVP basis set:
$DATA
Title
C1
H 1.0 -2.84267 1.60463 -0.00000
S 3
1 13.0107010 0.19682158E-01
2 1.9622572 0.13796524
3 0.44453796 0.47831935
S 1
1 0.12194962 1.0000000
P 1
1 0.8000000 1.0000000
H 1.0 -4.32187 0.58660 0.24007
S 3
1 13.0107010 0.19682158E-01
2 1.9622572 0.13796524
3 0.44453796 0.47831935
S 1
1 0.12194962 1.0000000
P 1
1 0.8000000 1.0000000
$ENDThree things to note here: 1) only the text after ‘HYDROGEN’ has to be taken from Basis Set Exchange, 2) After each atom, there has to be blank line, otherwise this will fail and 3) Basis sets have to be put in for all atoms, regardless of whether that same element was used before or not.
You can also use this method to input basis sets that are already in GAMESS. For example, I can use aug-cc-PVTZ for both H atoms:
$DATA
Title
C1
H 1.0 -2.84267 1.60463 -0.00000
ACCT
H 1.0 -4.32187 0.58660 0.24007
ACCT
$ENDAlso note that the $BASIS group must not be given with this.
- Inputting with BASNAM
In the group $BASIS there is a keyword called BASNAM. You can use it to input an array with names for every atom, and then define basis sets for particular names.
For example, For a methane molecule, I can name the first atom ‘atomc’ and the last three atoms ‘atomh’, and then define basis sets (3–21G) for them.
$BASIS BASNAM(1)=atomc,atomh,atomh,atomh $END
$DATA
Title
C1
C 6.0 -3.95207 1.60463 -0.00000
H 1.0 -2.84267 1.60463 -0.00000
H 1.0 -4.32187 0.58660 0.24007
H 1.0 -4.32187 1.90574 -1.00167
H 1.0 -4.32188 2.32155 0.76161
$END
$atomh
S 2
1 0.5447178000E+01 0.1562849787E+00
2 0.8245472400E+00 0.9046908767E+00
S 1
1 0.1831915800E+00 1.0000000
$end
$atomc
S 3
1 0.1722560000E+03 0.6176690738E-01
2 0.2591090000E+02 0.3587940429E+00
3 0.5533350000E+01 0.7007130837E+00
L 2
1 0.3664980000E+01 -0.3958951621E+00 0.2364599466E+00
2 0.7705450000E+00 0.1215834356E+01 0.8606188057E+00
L 1
1 0.1958570000E+00 0.1000000000E+01 0.1000000000E+01
$endThere are several things to note here — 1) The order of names in BASNAM will follow the order of the atoms in $DATA, so if you make a mistake in the order, the wrong basis set will be assigned to the wrong atom, 2) For inputting the basis set functions, all lines which have the exponents and coefficients (i.e. the lines that start with a number) have to be indented by one space (or one tab), 3) As in the case of $DATA, there has to be one blank line after each basis set input, 4) The names in BASNAM cannot match any of the keywords used in GAMESS, or any basis set names that are provided with GAMESS, and it has to be less than six characters, 5) Similar to $DATA, you can also use the basis sets that are provided with GAMESS.
In my example, I divided the atoms by element. You don’t have to do this, you can mix and match basis sets whatever way you want, as long as you follow the correct order of atoms.
- Inputting with BASNAM
GAMESS can also read the basis set information off a text file that is on the hard drive. This can be useful if you want to use a basis set that is not in GAMESS, but you don’t want to clutter the input file.
To use this, you have to put EXTFIL=.t. in the $BASIS section. Then put GBASIS=. This can be a bit tricky so I will explain with an example.
The input file should be like this:
$BASIS GBASIS=d2svp EXTFIL=.t. $END
$DATA
Title
C1
C 6.0 -6.48642 1.62990 0.00000
H 1.0 -5.41642 1.62990 0.00000
H 1.0 -6.84308 1.36449 -0.97327
H 1.0 -6.84309 2.60548 0.25678
H 1.0 -6.84309 0.91973 0.71649
$ENDEXTFIL=.t. requests that the basis set be read from an external file. Here, d2svp is the name of the basis set in the external file. There are a few caveats- 1)the basis set name defined in the external file cannot be more than 8 characters long, 2)the name can only have letters and numbers, 3)the name cannot match any of the basis sets already in the program i.e. names like ccd, cct cannot be used.
The external file has to be like this:
H d2svp
S 3
1 13.0107010 0.19682158E-01
2 1.9622572 0.13796524
3 0.44453796 0.47831935
S 1
1 0.12194962 1.0000000
P 1
1 0.8000000 1.0000000
He d2svp
S 3
1 38.354936737 0.23814288905E-01
2 5.7689081479 0.15490906777
3 1.2399407035 0.46998096633
S 1
1 0.29757815953 1.0000000
P 1
1 1.0000000 1.0000000(Only the top of the file is shown here)
The symbol of the element, and the name of that basis set has to be on the first line, then the basis set data, then a blank line before the next element. Note that GAMESS only supports shells with max. angular momentum 6 (i.e. I-orbitals).
Also, to allow the external file to be recognized, you have to edit the file rungms (rungms.bat for windows). The environment variable EXTBAS have to be set to the location of the .txt file containing the basis set. For Windows, the line should look like this:
@SET EXTBAS=%AUXDATADIR%\EXTBASIS.txtHere, I have put the EXTBASIS.txt file in the auxdata folder.
Since GAMESS does not contain a lot of the newer basis sets, and having them at hand is useful, I have put together a text file by extracting basis sets from another open-source QM program (psi4). This basis set file can be found here. Due to the 8-character constraint, I had to modify the names a little bit, all of that is explained on the github page. You can just download the file, and use it for your calculations. The basis sets are in GAMESS format, so will not work with other programs.
Using different basis sets on different atoms
You can use different basis sets on different atoms in your system. This is usually not recommended, as doing so may introduce artifacts in the result. Sometimes, it might be necessary to use a smaller basis set on hydrogen atoms (if they are just spectators and don’t participate in the reaction) to reduce the computational effort. In case of organometallic complexes, often the LANL2DZ(on metals) and 6–31+G* (on organic ligands) combination is used.
In GAMESS you can do this by putting them in $DATA or using BASNAM as I explained earlier.
GAMESS: https://www.msg.chem.iastate.edu/gamess/