This is the part 2 of the ongoing series of blogs, where I write about the how-to of the molecular modelling. These are focused mainly on its software side (GAMESS) instead of the theory.
Optimization is a process is that is routine in molecular modelling. It is generally used to find stationary points where the gradient of the energy is zero. These stationary points are energy minima, maxima which you would call reactants, products, transition states etc. (Usually optimization means finding the minima, but transition state search works on the same principle)
How does optimization work?
We can represent a molecule by the 3D cartesian coordinates of its atoms. If the QM program has a set of cartesian coordinates and atoms, it can approximate to the solution of the Schrödinger’s equation, which would give the total energy of that system. When any of the atoms are moved, that energy will change. Each atom can be moved in 3 directions (x,y,z) so we can express the energy of the system as a function of the cartesian coordinates of each of its atoms —
E=f(x_{1},y_{1},z_{1},x_{2},y_{2},z_{2},...)
Now, the optimized i.e. lowest energy structure would be the set of coordinates where E is minimum. That means the gradient (i.e. first derivative) of energy with respect to all coordinates has to be zero. Additionally, the second derivative has to be positive.
Optimization algorithms in quantum chemistry are influenced by the optimization method of Newton. Newton’s original method takes a initial set of coordinates and takes iterative steps to reach the stationary points. Each step requires the first and second derivatives of the E. As calculating second derivatives is costly, especially for bigger systems, there are so-called quasi-Newton algorithms that do not require accurate second-derivatives.
Most quantum chemistry programs including GAMESS, use the quasi methods. These methods guess the approximate second derivatives (often called guess hessian) but use accurate first derivatives to get to the lowest energy structure.
Choice of coordinate system
Writing the energy in terms of cartesian coordinates is not the only way. We can also write the energy in terms of chemically intuitive properties of the molecule like bond lengths(r), bond angles(α), torsional angles(β) etc.
E=f(l_{1},l_{2},...,\alpha_{1},\alpha_{2},...,\beta_{1},\beta_{2},...)
This coordinate system is called the internal coordinate system. Notice however, that the number of internal coordinates must be 3N-6 if N is the number of cartesian coordinates. In cartesian, there are 6 degrees of freedom(e.g. all atoms moving in x direction, all atoms rotating around an axis etc.) which represent the whole molecule moving in space, or rotating on an axis. These displacements do not change the energy in any way, and are eliminated in the internal coordinate system. (In case of linear molecules, there are 3N-5 internal coordinates, as only two rotational axes exist.)
There can be multiple choices of internal coordinates (any combination would work, as long as there are 3N-6 coordinates).
Using delocalised internal coordinates in GAMESS
There are multiple ways to use internal coordinates.
When Avogadro generates input for GAMESS, by default the molecule is represented in cartesian coordinates. In this case, you can set in $CONTRL, NZVAR=number of internal coordinates i.e. 3N-6 (or 3N-5, if linear). This requests the program to use internal coordinates for optimization.
$CONTRL SCFTYP=RHF RUNTYP=OPTIMIZE DFTTYP=B3LYP NZVAR=33 $END
$STATPT OPTTOL=0.0004 NSTEP=100 $END
$ZMAT DLC=.t. AUTO=.t. $ENDTop part of input file for benzene
You also have to add $ZMAT DLC=.t. AUTO=.t. $END. This instructs the program to automatically construct internal coordinates from cartesians. (The coordinates generated are a certain type called delocalised redundant internals) The program detects bonds by checking if the distance between two atoms is smaller than the sum of their van der Waals radii. This might fail if there are two atoms connected by a weak interaction, like a hydrogen bond.
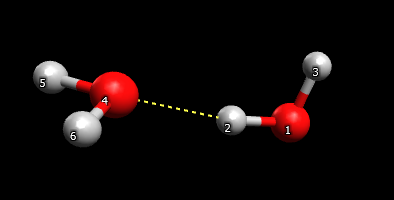
Two water molecules connected by an H-bond
In that case, the program needs to be told about the weak interaction by adding the keyword NONVDW to $ZMAT.
$BASIS GBASIS=N31 NGAUSS=6 NDFUNC=1 NPFUNC=1 DIFFSP=.TRUE. #END
$CONTRL SCFTYP=RHF RUNTYP=OPTIMIZE DFTTYP=B3LYP NZVAR=12 $END
$STATPT OPTTOL=0.0004 NSTEP=100 $END
$ZMAT DLC=.t. AUTO=.t. NONVDW(1)=2,4 $ENDThis keyword takes an array of numbers and reads two at a time: NONVDW(1)=i,j,k,l,m,n,…. this represents bonds between atom i and atom j, atom k and l, atom m and n and so on. (Hint: To see the labels in Avogadro, check the ‘label’ box on left panel, and then click the spanner, and change the ‘Text’ to ‘Atom number’.)
Using z-matrix input
Another way to use internal coordinates would be to supply them directly to GAMESS in the $DATA section. The internal coordinates are usually written in a specific format called the Z-matrix. Avogadro can generate the z-matrix, however, to get that you have to go through Extensions>Gaussian and then select ‘Z-matrix(compact)’ in the Format option.
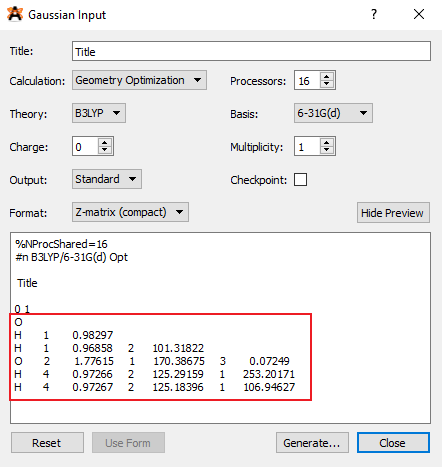
Z-matrix for water-water system
The z-matrix starts after the charge and multiplicity (0 1 in this case). Copy that text, and paste in into the $DATA section of the GAMESS input file.
$BASIS GBASIS=N31 NGAUSS=6 NDFUNC=1 NPFUNC=1 DIFFSP=.TRUE. $END
$CONTRL SCFTYP=RHF RUNTYP=OPTIMIZE DFTTYP=B3LYP COORD=ZMT
NZVAR=12 $END
$STATPT OPTTOL=0.0004 NSTEP=100 $END
$DATA
Title
C1
C 6.0 -5.80406 -1.22088 0.00000
O 8.0 -4.73406 -1.22088 0.00000
H 1.0 -6.16072 -0.32416 0.46215
H 1.0 -6.16073 -2.06948 0.54550
H 1.0 -6.16073 -1.26901 -1.00766
H 1.0 -4.41073 -1.17726 0.91348
$END
Don’t forget to put COORD=ZMT in $CONTRL so that GAMESS expects Gaussian style z-matrix input. Inputting NZVAR is also required to instruct the program to use internal coordinates. The benefit of this method is that you can edit and modify the internal coordinates yourself, if required.
There are also other methods for inputting various types of internal coordinates, but I am not mentioning them here.
Effect of coordinate choice on optimization
Using internal coordinates usually speeds up the optimization by a large extent. Consider the following benchmark for the water-water system (with B3LYP/6–31+G(d)):
| Cartesian coordinates | Delocalised internal coordinates | Z-matrix internal coordinates | |
| Number of optimization steps | 21 | 15 | 17 |
| Time taken (minutes) | 2.3 | 1.7 | 1.8 |
Optimization of water-water system
As you can see, auto generated internal coordinates are the best choice, and it can cut down the optimization time by a lot.
In case of molecules with rings, the z-matrix method does not work very well. In that case use the auto generation method, or if that fails, use cartesian. Also, there are some systems where the bond angles, or dihedral angles are 180 degrees. Internal coordinates can run into problems in those cases too.[ref-1]
The bottom line is that use cartesian coordinates as the last resort.
Reference-1: https://doi.org/10.1002/qua.560440821
GAMESS: https://www.msg.chem.iastate.edu/gamess/
Avogadro: https://avogadro.cc/